Ksenija Gasic, Stone Fruit Team Leader, Clemson University and Mercy Olmstead, Extension Team Leader, UF
One of the most serious diseases affecting peach production worldwide is bacterial spot, caused by Xanthomonas arboricola pv. pruni (Xap). Bacterial diseases are particularly difficult to control, because unlike infections in humans, which can be quickly quelled with antibiotics, in stone fruit antibacterial sprays are only effective in years with low to medium disease pressure.

Bacterial spot affects all stone fruit crops and can cause severe economic losses. Symptoms can appear on leaves, twigs, or fruit (Figure 1). Spots typically show up on leaves initially and as they grow larger they can become necrotic, giving a “shot-hole” appearance. Severe infestations can lead to premature defoliation, weakening the trees and depleting carbohydrate reserves. Lesions on fruit render it unmarketable, resulting in severe economic losses.
Traditional methods of control included antibacterial sprays such as oxytetracycline or copper-based compounds. However, concern with excessive antibiotic use, increasing heavy metals in the environment, and weak efficacy have led to growers to seek alternative control methods. Growers rely primarily on:
- Tested and certified nursery material (such as through the Clean Plant Network)
- Growing cultivars with a high degree of tolerance and/or resistance
- Breeding efforts to incorporate tolerance and/or resistance characteristics
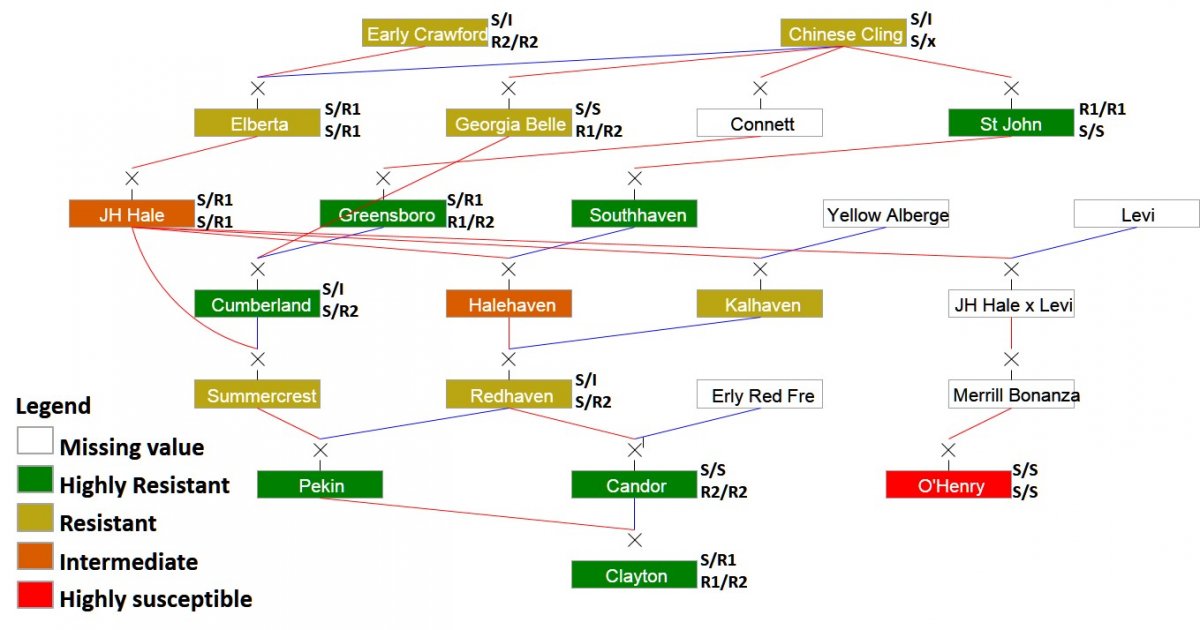
Thus, bacterial spot is a major focus for the peach breeding team in RosBREED, led by Ksenija Gasic. She has been working on the incorporation of bacterial spot tolerance and/or resistance into her breeding program at Clemson University. In addition, Dr. Gasic is attempting to find DNA markers that correlate with this tolerance and/or resistance. Previous research at North Carolina State University by Denny Werner and David Ritchie identified peach cultivars with exceptional tolerance and/or resistance, particularly in ‘Clayton’.
Cultivars such as ‘O’Henry’ introduced into the Southeast from California are most susceptible. Different genes are involved in the resistance for leaves and the fruit separately and together, but it was not known if resistance alleles from different genes were inherited separately or together in seedling progeny. However, as a result of the first RosBREED project, alleles associated with different levels of peach fruit resistance to bacterial spot were identified (Gasic et al., in press), and DNA tests with 80% accuracy in predicting fruit resistance are now available for routine screening.
The new project’s efforts of peach breeders Dr. Gasic (Clemson University) and Dr. Clark (University of Arkansas) together with the Pathology Team are focused on identifying and combining new genetic sources of both fruit and leaf resistance as well as developing reliable DNA tests for leaf resistance to bacterial spot for accurate and efficient evaluation of genetic potential in breeding germplasm.
References:
Okie WR. 1998. Handbook of Peach and Nectarine Varieties – Performance in the Southeastern United States and Index of Names. USDA/ARS Agric. Handbook 714.
Gasic K, Reighard G, Okie W, Clark J, Gradziel T, Byrne D, Peace C, Stegmeir T, Rosyara T and Iezzoni A. Bacterial spot resistance in peach: functional allele distribution in breeding germplasm. VIII International Peach Symposium, 17-20 June 2013, Matera, Italy, Acta Hort., 1084:69-74.
Voorrips RE, Bink MC, van de Weg WE. 2012. Pedimap: Software for visualization of genetic and phenotypic data in pedigrees. J Hered. 103(6):903-907.
